Before running any commands, be sure your virtual python environment is activated. Type activate pycgm39
Similarly to plot commands, emg commands propose convenient emg representation
To find out the different plot commands, type
pycgm2.exe NEXUS Plots EMG -h
Except temporal representation, all commands require you previously identified gait events
flowchart LR
id0[load a gait trial with model outputs]
id01[marked two gait trials \n in eclipse]
id02[marked *n* gait trials \n in eclipse]
subgraph EMG
subgraph emgSub[" "]
id41[temporal representation]
id411[pycgm2.exe NEXUS Plots EMG Temporal ]
style id411 fill:#000000,stroke:#333,stroke-width:4px,color:#ffffff
id42[Time-Normalized representation]
id421[pycgm2.exe NEXUS Plots EMG Normalized ]
style id421 fill:#000000,stroke:#333,stroke-width:4px,color:#ffffff
end
id43[Comparison]
id431[pycgm2.exe NEXUS Plots EMG Comparison ]
style id431 fill:#000000,stroke:#333,stroke-width:4px,color:#ffffff
end
id0--->emgSub
id41--command---id411
id42--command---id421
id43--command---id431
id01---> id43
id02---> id42
To know input arguments
- type
pycgm2.exe NEXUS Plots EMG Normalized -h - Refer to the documentation API
Preliminary settings
EMG plot command failed if you have not labelled your analog channels as follow
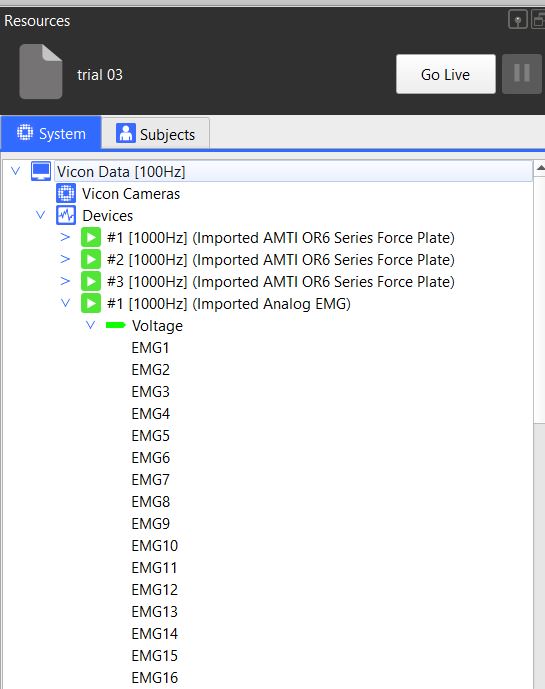
We predefined the location of 16 emg devices.
| EMG channel | Muscle | Side |
|---|---|---|
| EMG1 | rectus femoris | Left |
| EMG2 | rectus femoris | Right |
| EMG3 | vastus lateralis | Left |
| EMG4 | vastus lateralis | Right |
| EMG5 | semitendinosus | Left |
| EMG6 | semitendinosus | Right |
| EMG7 | tibialis anterior | Left |
| EMG8 | tibialis anterior | Right |
| EMG9 | soleus | Left |
| EMG10 | soleus | Right |
| EMG11 | None | None |
| EMG12 | None | None |
| EMG13 | None | None |
| EMG14 | None | None |
| EMG15 | None | None |
| EMG16 | None | None |
Consult the FAQ page if you want to modify this configuration.